Information Processing by cells
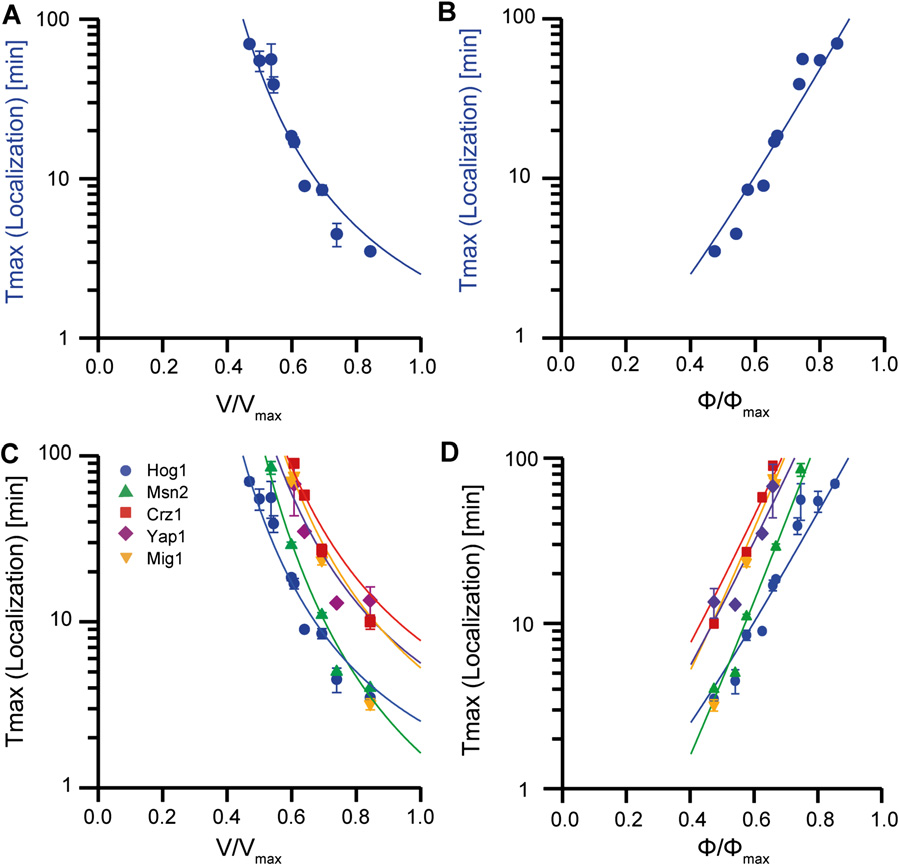
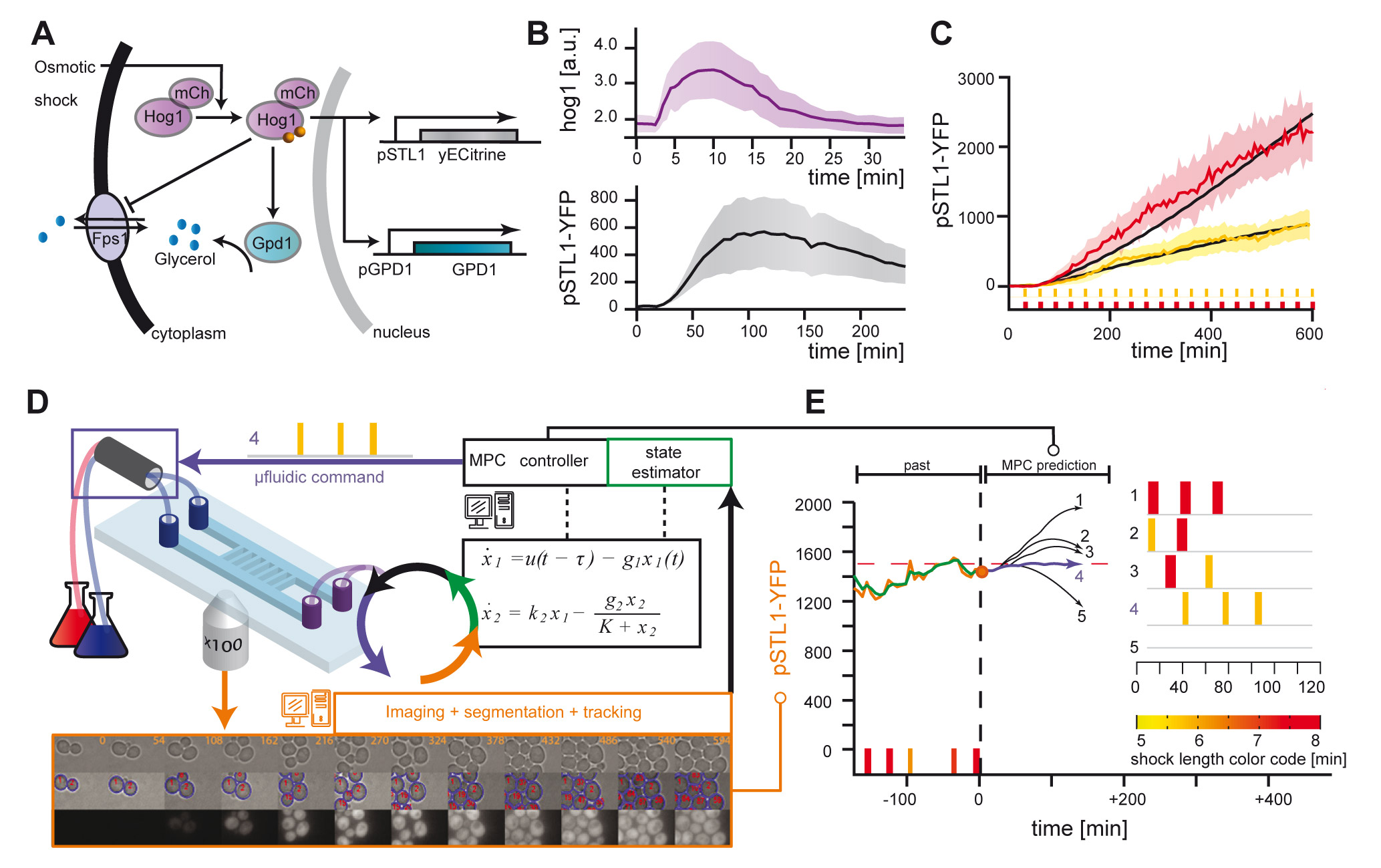
Cells have evolved complex signaling pathways to cope with environmental stress. Quantitative biology and interdisciplinary approaches can be used to probe the dynamics of signaling pathways in living cells. We studied the signal processing ability of yeast cells in response to osmotic stress ...[+]
Physical Limits of cell signaling
Whereas the genetic background is a key regulator of cellular functions, the physico chemical environment has long been considered as a major regulator of global cellular functions. Cells behaviours depend on temperature, pH, light... We show here that molecular crowding may be a major regulator of biochemical rates inside the cells... [+]
Cybergenetics: Real Time Control of gene expression
The development of cell - computer interfaces is very recent and yet bear fantastic potential applications. We are among the first to set up an experimentla platform for the real time control of gene expression in yeast at the single cell level... [+]
Synthetic biology and, more generally, genome editing are fantastic tools and concepts for engineers who wants to explore, tinker, hack the functioning of living systems. Using and developping tools of genome editing is part of our primary research activities...[+]
Genetic drift is a passive selection process, usually at play in small population, where the probability of fixation of a neutral mutation is non null. We are interested in how this important effect in population dynamics is also at play in large, spatially structured population of yeast and bacteria... [+]
Ideal Cylindrical Colonies
We design a novel method to grow yeast and bacteria colonies into any arbitrary geometries. We are studying how efficiently cell assemblies grow when their shape is controlled. In particular we are studying cylindrical colony that are a perfect candidate for relating the metabolic yield to the growth of the population... [+]
C. elegans, a 1 mm long worm, displays several interesting behavior, that are regulated by its simple, yet efficient, neural network. It is able to forage for food, respond to mechanical stimulus, adapt its locomotion pattern from crawling to swimming. Importantly, it can feel the presence of electric field. We are studying how aging and locomotion using electrotaxis to control worms episode of locomotion...[+]
C. elegans is a small worm which moves by propagating a flexural wave along its body. We are studying how it adapts its locomotion pattern to its mechanical environment. Worms locomotion is indeed passively modulated by the resistance of its environment and possibly through proprioception...[+]
Mamalian cells are known to both exert forces on their substrate and to react physiologically to forces exerted on them. We have studied various aspects of force generation and collective cell migration using microfabrication methods. We are now imaging with super resolution microscopy the structure of focal adhesion complexes, which are a key unit in mechano sensing and force transmission between the cytoskeleton and the extra cellular matrix... [+]
Cell Based Assays are an interesting alternative to biochemical assays performed on live animals. The first advantage is to be able to screen for drugs and measure quantitatively the toxicity of chemicals directly at the single cell level...[+]
Sand dunes can adopt various shapes depending on the directionality of the local winds and the sand supply. The most simple, and studied dune is the barchan, a crescent shape dune that forms in desert blown by very directional wind. We were the first to develop a laboratory experiment to study downsized version of these structure in the lab, opening the way to several key findings on the dynamics of barchan dunes...[+]
Barchan dunes are crescent shaped sand dunes that migrate downwind. The smaller they are, the faster. To get an order of magnitude, sand dunes can go up to several tens of meters a year in aeolian desert for the smallest ones. Since not all dunes have the same size, they travel at different speed and can collide. The result of such events is shown to be a key regulator of the structure of deserts...[+]
Sand dunes can adopt the shape of long ridges which orient either transversally or longitudinally to the average wind direction. Such shapes form primarily when the wind direction alternates between two dominant orientations. We upgraded our experimental setup to study the stability and formation of such dunes...[+]
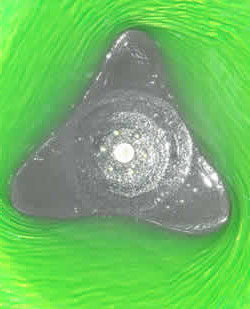
It's well known that the surface of a liquid in a rotating cylinder will adopt a paraboloid like shape. Strikingly this is not always the case, and we observed that polygonal shapes can appear at the surface of the liquid...[+]